 Um trabalho de pós-doutorando do Departamento de Fitopatologia, Dr. Miller Lehner, ex-integrante do Laboratório de Biologia de Populações (Prof. Mizubuti), acaba de ser publicado na revista PLoS ONE. O estudo foi conduzido em parceria com o Laboratório da Profa. Sarah Pethybridge (Cornell University) onde o pesquisador atuou como pós-doc associado no ano de 2016, após atuar no DFP.
Um trabalho de pós-doutorando do Departamento de Fitopatologia, Dr. Miller Lehner, ex-integrante do Laboratório de Biologia de Populações (Prof. Mizubuti), acaba de ser publicado na revista PLoS ONE. O estudo foi conduzido em parceria com o Laboratório da Profa. Sarah Pethybridge (Cornell University) onde o pesquisador atuou como pós-doc associado no ano de 2016, após atuar no DFP.
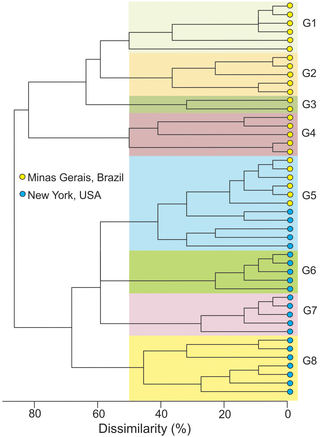
No estudo foram comparadas duas populações de Sclerotinia sclerotiorum, agente causal da doença conhecida como podridão de Sclerotinia ou mofo-branco, quanto a variabilidade genética do fungo. Pela primeira vez são comparadas diretamente populações de uma região tropical (Minas Gerais) com populações de regiões de clima temperado (Estado de Nova York), cujos níveis de variabilidade genética, inferidos por marcadores microsatélites, não parecem ter ser influenciados pelo clima predominante na região de origem. O Dr. Lehner atualmente é pesquisador na Monsanto do Brasil.
Resumo – Clique aqui para ler o trabalho na íntegra
Sclerotinia sclerotiorum populations from tropical agricultural zones have been suggested to be more variable compared to those from temperate zones. However, no data were available comparing populations from both zones using the same set of markers. In this study, we compared S. sclerotiorum populations from the United States of America (USA, temperate) and southeast Brazil (tropical) using the frequency of mycelial compatibility groups (MCGs) and 13 microsatellite (SSR) markers. Populations were sourced from diseased plants within leguminous crops in New York, USA (NY; n = 78 isolates), and Minas Gerais State, Brazil (MG; n = 109). Twenty MCGs were identified in NY and 14 were previously reported in MG. The effective number of genotypes based on Hill’s number of order 0, which corresponded to the number of multilocus genotypes (MLGs) were 22 (95% CI = 15.6–28.4) and 24 (95% CI = 18.9–29.1) in NY and MG, respectively. Clonal fractions of MLGs were 71.8% (NY) and 78.0% (MG). The effective number of genotypes based on Hill’s number of orders 1 and 2 in NY were 8.9 (95% CI = 5.2–12.6) and 4.4 (95% CI = 2.6–6.1), respectively. For MG these indices were 11.4 (95% CI = 8.7–14.1) and 7.1 (95% CI = 5.1–9.0), respectively. There were no significant differences of allelic richness, private allelic richness, gene diversity, effective number of alleles and genotype evenness between the NY and MG populations. The populations were differentiated, with 29% of total variance attributed to differences between them and G”ST and Jost’s D indices higher than 0.50. Cluster analysis revealed dissimilarity higher than 80% among most MLGs from both populations. Different alleles segregated in the populations but both had similar levels of genotypic variability.
